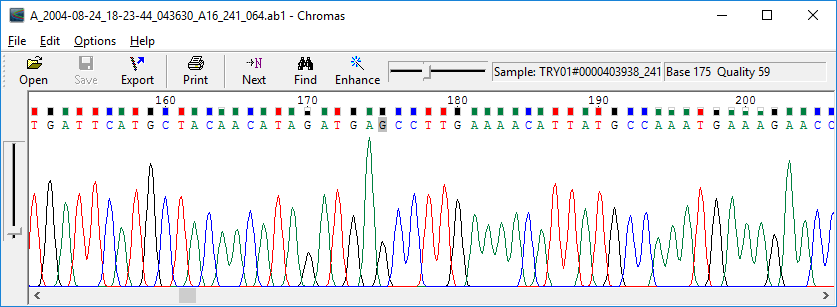
Install the ab1 file viewer. There are several free software tools. For Mac OS X, use 4Peaks tool. Fox MS Windows, use chromas tools. You need a sequence alignment tool for 16S gene sequences. We recommend the EzEditor2 tool Tutorial of EzEditor2. The following example ab1 files are required. I'm looking for a software (free or paid) that can merge Sanger sequences from the same gene, automatically determine the best nucleotide based on quality (if overlapping) and ideally blast.
- Sanger Sequence Assembler software, free download Macos 7
- Sanger Sequence Assembler software, free download Macos Windows 7
- Sanger Sequence Assembler software, free download Macos Download
- Sanger Sequence Assembler software, free download Macos Windows 10
Sanger sequencing is a method of DNA sequencing based on the selective incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitroDNA replication.[1][2] After first being developed by Frederick Sanger and colleagues in 1977, it became the most widely used sequencing method for approximately 40 years. It was first commercialized by Applied Biosystems in 1986.[3] More recently, higher volume Sanger sequencing has been replaced by 'Next-Gen' sequencing methods, especially for large-scale, automated genome analyses. However, the Sanger method remains in wide use, for smaller-scale projects, and for validation of Next-Gen results. It still has the advantage over short-read sequencing technologies (like Illumina) in that it can produce DNA sequence reads of > 500 nucleotides.
Sanger Sequence Assembler software, free download Macos 7
Functions to analyse sanger sequencing reads in R. Contribute to roblanf/sangeranalyseR development by creating an account on GitHub. ACME is a free cross assembler released under the GNU GPL. It can produce code for the following processors: 6502, 6510 (including illegal opcodes), 65c02 and 65816. ACME supports the standard assembler stuff like global/local/anonymous labels, offset assembly, conditional assembly and looping assembly. It can include other source files as well as binaries while assembling.
Method[edit]
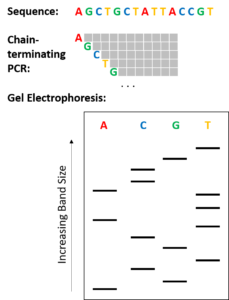
The classical chain-termination method requires a single-stranded DNA template, a DNA primer, a DNA polymerase, normal deoxynucleotidetriphosphates (dNTPs), and modified di-deoxynucleotidetriphosphates (ddNTPs), the latter of which terminate DNA strand elongation. These chain-terminating nucleotides lack a 3'-OH group required for the formation of a phosphodiester bond between two nucleotides, causing DNA polymerase to cease extension of DNA when a modified ddNTP is incorporated. The ddNTPs may be radioactively or fluorescently labelled for detection in automated sequencing machines.
The DNA sample is divided into four separate sequencing reactions, containing all four of the standard deoxynucleotides (dATP, dGTP, dCTP and dTTP) and the DNA polymerase. To each reaction is added only one of the four dideoxynucleotides (ddATP, ddGTP, ddCTP, or ddTTP), while the other added nucleotides are ordinary ones. The dideoxynucleotide concentration should be approximately 100-fold higher than that of the corresponding deoxynucleotide (e.g. 0.5mM ddTTP : 0.005mM dTTP) to allow enough fragments to be produced while still transcribing the complete sequence(but the concentration of ddNTP also depends on the desired length of sequence).[2] Putting it in a more sensible order, four separate reactions are needed in this process to test all four ddNTPs. Following rounds of template DNA extension from the bound primer, the resulting DNA fragments are heat denatured and separated by size using gel electrophoresis. In the original publication of 1977,[2] the formation of base-paired loops of ssDNA was a cause of serious difficulty in resolving bands at some locations. This is frequently performed using a denaturing polyacrylamide-urea gel with each of the four reactions run in one of four individual lanes (lanes A, T, G, C). The DNA bands may then be visualized by autoradiography or UV light and the DNA sequence can be directly read off the X-ray film or gel image.
In the image on the right, X-ray film was exposed to the gel, and the dark bands correspond to DNA fragments of different lengths. A dark band in a lane indicates a DNA fragment that is the result of chain termination after incorporation of a dideoxynucleotide (ddATP, ddGTP, ddCTP, or ddTTP). The relative positions of the different bands among the four lanes, from bottom to top, are then used to read the DNA sequence.
Technical variations of chain-termination sequencing include tagging with nucleotides containing radioactive phosphorus for radiolabelling, or using a primer labeled at the 5' end with a fluorescent dye. Dye-primer sequencing facilitates reading in an optical system for faster and more economical analysis and automation. The later development by Leroy Hood and coworkers[4][5] of fluorescently labeled ddNTPs and primers set the stage for automated, high-throughput DNA sequencing.
Chain-termination methods have greatly simplified DNA sequencing. For example, chain-termination-based kits are commercially available that contain the reagents needed for sequencing, pre-aliquoted and ready to use. Limitations include non-specific binding of the primer to the DNA, affecting accurate read-out of the DNA sequence, and DNA secondary structures affecting the fidelity of the sequence.
Dye-terminator sequencing[edit]
Dye-terminator sequencing utilizes labelling of the chain terminator ddNTPs, which permits sequencing in a single reaction, rather than four reactions as in the labelled-primer method. In dye-terminator sequencing, each of the four dideoxynucleotide chain terminators is labelled with fluorescent dyes, each of which emit light at different wavelengths.
Sanger Sequence Assembler software, free download Macos Windows 7
Owing to its greater expediency and speed, dye-terminator sequencing is now the mainstay in automated sequencing. Its limitations include dye effects due to differences in the incorporation of the dye-labelled chain terminators into the DNA fragment, resulting in unequal peak heights and shapes in the electronic DNA sequence trace chromatogram after capillary electrophoresis (see figure to the left).
This problem has been addressed with the use of modified DNA polymerase enzyme systems and dyes that minimize incorporation variability, as well as methods for eliminating 'dye blobs'. The dye-terminator sequencing method, along with automated high-throughput DNA sequence analyzers, was used for the vast majority of sequencing projects until the introduction of Next Generation Sequencing.
Automation and sample preparation[edit]
Sanger Sequence Assembler software, free download Macos Download
Automated DNA-sequencing instruments (DNA sequencers) can sequence up to 384 DNA samples in a single batch. Batch runs may occur up to 24 times a day. DNA sequencers separate strands by size (or length) using capillary electrophoresis, they detect and record dye fluorescence, and output data as fluorescent peak trace chromatograms. Sequencing reactions (thermocycling and labelling), cleanup and re-suspension of samples in a buffer solution are performed separately, before loading samples onto the sequencer. A number of commercial and non-commercial software packages can trim low-quality DNA traces automatically. These programs score the quality of each peak and remove low-quality base peaks (which are generally located at the ends of the sequence). The accuracy of such algorithms is inferior to visual examination by a human operator, but is adequate for automated processing of large sequence data sets.
Challenges[edit]
Common challenges of DNA sequencing with the Sanger method include poor quality in the first 15-40 bases of the sequence due to primer binding[citation needed] and deteriorating quality of sequencing traces after 700-900 bases. Base calling software such as Phred typically provides an estimate of quality to aid in trimming of low-quality regions of sequences.[6][7]
In cases where DNA fragments are cloned before sequencing, the resulting sequence may contain parts of the cloning vector. In contrast, PCR-based cloning and next-generation sequencing technologies based on pyrosequencing often avoid using cloning vectors. Recently, one-step Sanger sequencing (combined amplification and sequencing) methods such as Ampliseq and SeqSharp have been developed that allow rapid sequencing of target genes without cloning or prior amplification.[8][9]
Current methods can directly sequence only relatively short (300-1000 nucleotides long) DNA fragments in a single reaction. The main obstacle to sequencing DNA fragments above this size limit is insufficient power of separation for resolving large DNA fragments that differ in length by only one nucleotide.
Microfluidic Sanger sequencing[edit]
Microfluidic Sanger sequencing is a lab-on-a-chip application for DNA sequencing, in which the Sanger sequencing steps (thermal cycling, sample purification, and capillary electrophoresis) are integrated on a wafer-scale chip using nanoliter-scale sample volumes. This technology generates long and accurate sequence reads, while obviating many of the significant shortcomings of the conventional Sanger method (e.g. high consumption of expensive reagents, reliance on expensive equipment, personnel-intensive manipulations, etc.) by integrating and automating the Sanger sequencing steps.
In its modern inception, high-throughput genome sequencing involves fragmenting the genome into small single-stranded pieces, followed by amplification of the fragments by Polymerase Chain Reaction (PCR). Adopting the Sanger method, each DNA fragment is irreversibly terminated with the incorporation of a fluorescently labeled dideoxy chain-terminating nucleotide, thereby producing a DNA “ladder” of fragments that each differ in length by one base and bear a base-specific fluorescent label at the terminal base. Amplified base ladders are then separated by Capillary Array Electrophoresis (CAE) with automated, in situ “finish-line” detection of the fluorescently labeled ssDNA fragments, which provides an ordered sequence of the fragments. These sequence reads are then computer assembled into overlapping or contiguous sequences (termed 'contigs') which resemble the full genomic sequence once fully assembled.[10]
Sanger methods achieve read lengths of approximately 800bp (typically 500-600bp with non-enriched DNA). The longer read lengths in Sanger methods display significant advantages over other sequencing methods especially in terms of sequencing repetitive regions of the genome. A challenge of short-read sequence data is particularly an issue in sequencing new genomes (de novo) and in sequencing highly rearranged genome segments, typically those seen of cancer genomes or in regions of chromosomes that exhibit structural variation.[11]
Applications of microfluidic sequencing technologies[edit]
Other useful applications of DNA sequencing include single nucleotide polymorphism (SNP) detection, single-strand conformation polymorphism (SSCP) heteroduplex analysis, and short tandem repeat (STR) analysis. Resolving DNA fragments according to differences in size and/or conformation is the most critical step in studying these features of the genome.[10]
Device design[edit]
The sequencing chip has a four-layer construction, consisting of three 100-mm-diameter glass wafers (on which device elements are microfabricated) and a polydimethylsiloxane (PDMS) membrane. Reaction chambers and capillary electrophoresis channels are etched between the top two glass wafers, which are thermally bonded. Three-dimensional channel interconnections and microvalves are formed by the PDMS and bottom manifold glass wafer.
The device consists of three functional units, each corresponding to the Sanger sequencing steps. The Thermal Cycling (TC) unit is a 250-nanoliter reaction chamber with integrated resistive temperature detector, microvalves, and a surface heater. Movement of reagent between the top all-glass layer and the lower glass-PDMS layer occurs through 500-μm-diameter via-holes. After thermal-cycling, the reaction mixture undergoes purification in the capture/purification chamber, and then is injected into the capillary electrophoresis (CE) chamber. The CE unit consists of a 30-cm capillary which is folded into a compact switchback pattern via 65-μm-wide turns.
Sequencing chemistry[edit]
- Thermal cycling
- In the TC reaction chamber, dye-terminator sequencing reagent, template DNA, and primers are loaded into the TC chamber and thermal-cycled for 35 cycles ( at 95 °C for 12 seconds and at 60 °C for 55 seconds).
- Purification
- The charged reaction mixture (containing extension fragments, template DNA, and excess sequencing reagent) is conducted through a capture/purification chamber at 30 °C via a 33-Volts/cm electric field applied between capture outlet and inlet ports. The capture gel through which the sample is driven, consists of 40 μM of oligonucleotide (complementary to the primers) covalently bound to a polyacrylamide matrix. Extension fragments are immobilized by the gel matrix, and excess primer, template, free nucleotides, and salts are eluted through the capture waste port. The capture gel is heated to 67-75 °C to release extension fragments.
- Capillary electrophoresis
- Extension fragments are injected into the CE chamber where they are electrophoresed through a 125-167-V/cm field.
Platforms[edit]
The Apollo 100 platform (Microchip Biotechnologies Inc., Dublin, CA)[12] integrates the first two Sanger sequencing steps (thermal cycling and purification) in a fully automated system. The manufacturer claims that samples are ready for capillary electrophoresis within three hours of the sample and reagents being loaded into the system. The Apollo 100 platform requires sub-microliter volumes of reagents.
Comparisons to other sequencing techniques[edit]
| Technology | Number of lanes | Injection volume (nL) | Analysis time | Average read length | Throughput (including analysis; Mb/h) | Gel pouring | Lane tracking |
|---|---|---|---|---|---|---|---|
| Slab gel | 96 | 500–1000 | 6–8 hours | 700 bp | 0.0672 | Yes | Yes |
| Capillary array electrophoresis | 96 | 1–5 | 1–3 hours | 700 bp | 0.166 | No | No |
| Microchip | 96 | 0.1–0.5 | 6–30 minutes | 430 bp | 0.660 | No | No |
| 454/Roche FLX (2008) | < 0.001 | 4 hours | 200–300 bp | 20–30 | |||
| Illumina/Solexa (2008) | 2–3 days | 30–100 bp | 20 | ||||
| ABI/SOLiD (2008) | 8 days | 35 bp | 5–15 | ||||
| Illumina MiSeq (2019) | 1–3 days | 2x75–2x300 bp | 170–250 | ||||
| Illumina NovaSeq (2019) | 1–2 days | 2x50–2x150 bp | 22,000–67,000 | ||||
| Ion Torrent Ion 530 (2019) | 2.5–4 hours | 200–600 bp | 110–920 | ||||
| BGI MGISEQ-T7 (2019) | 1 day | 2x150 bp | 250,000 | ||||
| Pacific Biosciences (2019) | 10–20 hours | 10–30 kb | 1,300 | ||||
| Oxford Nanopore MinIon (2019) | 3 days | 13–20 kb[15] | 700 |
Sanger Sequence Assembler software, free download Macos Windows 10
The ultimate goal of high-throughput sequencing is to develop systems that are low-cost, and extremely efficient at obtaining extended (longer) read lengths. Longer read lengths of each single electrophoretic separation, substantially reduces the cost associated with de novo DNA sequencing and the number of templates needed to sequence DNA contigs at a given redundancy. Microfluidics may allow for faster, cheaper and easier sequence assembly.[10]
References[edit]
- ^Sanger F; Coulson AR (May 1975). 'A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase'. J. Mol. Biol. 94 (3): 441–8. doi:10.1016/0022-2836(75)90213-2. PMID1100841.
- ^ abcSanger F; Nicklen S; Coulson AR (December 1977). 'DNA sequencing with chain-terminating inhibitors'. Proc. Natl. Acad. Sci. U.S.A. 74 (12): 5463–7. Bibcode:1977PNAS..74.5463S. doi:10.1073/pnas.74.12.5463. PMC431765. PMID271968.
- ^Adams, Jill U. (2008). 'DNA Sequencing Technologies'. Nature Education. Retrieved October 24, 2019.
- ^Smith LM, Sanders JZ, Kaiser RJ, et al. (1986). 'Fluorescence detection in automated DNA sequence analysis'. Nature. 321 (6071): 674–9. Bibcode:1986Natur.321.674S. doi:10.1038/321674a0. PMID3713851.
We have developed a method for the partial automation of DNA sequence analysis. Fluorescence detection of the DNA fragments is accomplished by means of a fluorophore covalently attached to the oligonucleotide primer used in enzymatic DNA sequence analysis. A different coloured fluorophore is used for each of the reactions specific for the bases A, C, G and T. The reaction mixtures are combined and co-electrophoresed down a single polyacrylamide gel tube, the separated fluorescent bands of DNA are detected near the bottom of the tube, and the sequence information is acquired directly by computer.
- ^Smith LM; Fung S; Hunkapiller MW; Hunkapiller TJ; Hood LE (April 1985). 'The synthesis of oligonucleotides containing an aliphatic amino group at the 5' terminus: synthesis of fluorescent DNA primers for use in DNA sequence analysis'. Nucleic Acids Res. 13 (7): 2399–412. doi:10.1093/nar/13.7.2399. PMC341163. PMID4000959.
- ^'Phred - Quality Base Calling'. Retrieved 2011-02-24.
- ^Ledergerber, C; Dessimoz, C (2011). 'Base-calling for next-generation sequencing platforms'. Briefings in Bioinformatics. 12 (5): 489–97. doi:10.1093/bib/bbq077. PMC3178052. PMID21245079.
- ^Murphy, K.; Berg, K.; Eshleman, J. (2005). 'Sequencing of genomic DNA by combined amplification and cycle sequencing reaction'. Clinical Chemistry. 51 (1): 35–39. doi:10.1373/clinchem.2004.039164. PMID15514094.
- ^Sengupta, D. .; Cookson, B. . (2010). 'SeqSharp: A general approach for improving cycle-sequencing that facilitates a robust one-step combined amplification and sequencing method'. The Journal of Molecular Diagnostics. 12 (3): 272–277. doi:10.2353/jmoldx.2010.090134. PMC2860461. PMID20203000.
- ^ abcKan, Cheuk-Wai; Fredlake, Christopher P.; Doherty, Erin A. S.; Barron, Annelise E. (1 November 2004). 'DNA sequencing and genotyping in miniaturized electrophoresis systems'. Electrophoresis. 25 (21–22): 3564–3588. doi:10.1002/elps.200406161. PMID15565709.
- ^ abMorozova, Olena; Marra, Marco A (2008). 'Applications of next-generation sequencing technologies in functional genomics'. Genomics. 92 (5): 255–64. doi:10.1016/j.ygeno.2008.07.001. PMID18703132.
- ^Microchip Biologies Inc. Apollo 100
- ^Sinville, Rondedrick; Soper, Steven A (2007). 'High resolution DNA separations using microchip electrophoresis'. Journal of Separation Science. 30 (11): 1714–28. doi:10.1002/jssc.200700150. PMID17623451.
- ^Kumar, Kishore; Cowley, Mark; Davis, Ryan (2019). 'Next-Generation Sequencing and Emerging Technologies'. Seminars in Thrombosis and Hemostasis. 45 (7): 661–673. doi:10.1055/s-0039-1688446. ISSN0094-6176. PMID31096307.
- ^Tyson, John R.; O'Neil, Nigel J.; Jain, Miten; Olsen, Hugh E.; Hieter, Philip; Snutch, Terrance P. (2018). 'MinION-based long-read sequencing and assembly extends the Caenorhabditis elegans reference genome'. Genome Research. 28 (2): 266–274. doi:10.1101/gr.221184.117. ISSN1088-9051. PMC5793790. PMID29273626.
Further reading[edit]
- https://web.archive.org/web/20120214053015/http://nano.cancer.gov/news_center/nanotech_news_2006-05-30a.asp[full citation needed]
- https://web.archive.org/web/20120214053039/http://nano.cancer.gov/news_center/monthly_feature_2005_aug.asp[full citation needed]
- Dewey, F. E; Pan, S; Wheeler, M. T; Quake, S. R; Ashley, E. A (2012). 'DNA Sequencing: Clinical Applications of New DNA Sequencing Technologies'. Circulation. 125 (7): 931–44. doi:10.1161/CIRCULATIONAHA.110.972828. PMC3364518. PMID22354974.
- Sanger, F; Coulson, A.R; Barrell, B.G; Smith, A.J.H; Roe, B.A (1980). 'Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing'. Journal of Molecular Biology. 143 (2): 161–78. doi:10.1016/0022-2836(80)90196-5. PMID6260957.
External links[edit]
Are you music enthusiast? Ever wondered how well it would be if the music you here is supplied with all the additional information as titles, artists, albums and genres? Following is the list of software which offers this basic functionality with sequencing your huge music library and some of the rich and unique features. You can expect some free sequencer software while some are paid sequencer software. The list has some of the best-picked software of this category.
Related:
Mp3 Tag Pro
Mp3TagPro is free music sequencer software available for free download and use. This software is a powerful tool for all music formats. The software creates new subfolders and organizes the music files into them according to your pattern. The software will create folders with artists, genres and many other such categories at your one click.
MIDI Sequencer Software
MIDI sequencer software called Sekaiju is free, open source software for creating and editing MIDI data. MIDI stands for Multi Documented format. The software features unlimited redo and undo, 16 MIDI input ports and 16 MIDI ports all of them can be used simultaneously and much more.
Audio Sequencer Software
This software from Jazz MIDI sequencer is open source software under GNU GPL license. The Jazz++ is an excellent application for recording and mixing MIDI sequences. This software is actively supported for the Windows and Mac platforms.
Sequencer for Mac
Sequencer for Mac is the music sequencer software for Mac platform users. The software has a range of features to offer audio and MIDI recording, audio workstation, step sequencing, audio multitrack editor and much more. The software is available free of cost for download and use.
Sequencer for Windows
Sequencer software for Windows is free music sequencer software for Windows platform users. The free version features variable track lengths supporting 16 to 128 steps, three FX chains, 30 to 180 BPM tempo, adjustable pitch, pan, volume, and synth note length per step.
Caustic 3 for Android
Caustic 3 is the music sequencer software for Android platform users. You can create your rack by adding up to 14 machines from the various synthesizers this software has to offer. The software comes in demo free version to give you a taste of the software.
Anvil Studio – Most Popular Software
Anvil studio is the most popular software in the category of music sequencer software. The long list of features of this software includes recording music with MIDI and audio equipment, sequence music with MIDI equipment and much more. The software has a free of cost version for download and use. You can also see Audio mixer software
How to Install Music Sequencer Software?
Most of the above-listed music sequencer software offer installation file, which you can download and run the installation procedure to get the software installed. Music sequencer software is the software for sequencing your long list of music files, according to the artists, genres, and much more such categories. You can also see Guitar Recording Software
Use apple configurator 2 to backup ios apps to mac. Apple Configurator 1 will run on El Capitan but to take advantage of new features in iOS 9 you will need Apple Configurator 2. Please be aware that this is a new app, in fact Apple are pushing it as a different product to Apple Configurator 1. On first launch it will attempt to migrate your old settings across. Nov 04, 2015 All I'm armed with is a Mac with AC2. No MDM (at this point, we have Meraki, but I don't have the credentials yet to be able to use their mdm), no Apple Profile Configurator. All I want to do is be able to add apps to the iPads and keep them all uniformly configured. We have folders for each grade with the apps that they use. Aug 21, 2019 You can get more information from our Apple Configurator 2 guide. How to update your iOS devices using Apple Configurator 2. In order to update your iOS device you'll need to have it physically connected via USB to the Mac running Apple Configurator2. You can use a USB hub for multiple devices or if you have multiple daisy-chained displays, you. Apple Configurator 2 will backup your iOS device — but it ignores the apps. There is a solution: third-party app iMazing will let you manage your new and old apps on your Mac. Apple Configurator 2 makes it easy to deploy iPad, iPhone, iPod touch, and Apple TV devices in your institution. Use Apple Configurator to configure your devices. You can use Apple Configurator to quickly configure large numbers of devices with the settings, apps, and data you specify for your students, employees, or customers.
Almost all the above-listed software offers the basic functionality of sequencing your music files according to various categories. You can opt for the paid ones if you want fast and other advanced options, for edit and modify the music file you want.
The Economy Infographics Solution is available at $25US to users of ConceptDraw DIAGRAM v13 and ConceptDraw OFFICE v6, via STORE.ConceptDraw DIAGRAM is well-known for its ability to exchange documents with Microsoft Visio. The new solution provides all of the graphical user elements needed to transform economic and financial information into a compelling visual message.The Economy Infographics Solution contains 6 vector libraries composed of 316 scalable icons. Infographics software mac os x. It can open and save documents that can be used by Visio (VSD, VDX and VSDX documents) users. One can use this solution to quickly create eye catching and impressive infographics which can be used to effectively communicate economic and financial messages at a glance.